
Service Details
|
Service Name |
Length (nt) |
Purification |
Price/ |
Deliverable |
Application |
|
Common Oligos |
5-60 |
DSL/OPC/PAGE/HPLC |
Inquire |
· Tube or Customized · Lyophilized DNA · COA Report |
PCR、DNA Sequencing |
|
Long Oligos |
60+ |
PAGE/HPLC/Dual PAGE & HPLC |
NGS, genomic Research |
||
|
Large-scale Oligos |
Customized |
Customized |
New drug screening, drug production |
||
|
Modified Oligos |
5-120 |
DSL/OPC/PAGE/HPLC/ Dual PAGE & HPLC |
Various molecular biology research |
|
Modification List |
Degenerate Bases |
|||
|
5' Mod |
3' Mod |
Double Label Modifications |
Internal Modifications |
Codes |
|
5'Alexa Flour 350 |
3'Alexa Flour 350 |
CY3,BHQ2 |
(dT-Alexa Flour 350) | B=G,C,T |
|
5'Alexa Flour 405 |
3'Alexa Flour 405 |
CY3,MGB |
(dT-Alexa Flour 405) | D=A,G,T |
|
5'Alexa Flour 488 |
3'Alexa Flour 488 |
CY5,BHQ2 |
(dT-Alexa Flour 488) | H=A,C,T |
|
5'Alexa Flour 532 |
3'Alexa Flour 532 |
CY5,BHQ3 |
(dT-Alexa Flour 532) | K=G,T |
|
5'Alexa Flour 546 |
3'Alexa Flour 546 |
CY5,MGB |
(dT-Alexa Flour 546) | M=A,C |
|
5'Alexa Flour 555 |
3'Alexa Flour 555 |
CY5.5,BHQ2 |
(dT-Alexa Flour 555) | N=A,G,C,T |
|
5'Alexa Flour 568 |
3'Alexa Flour 568 |
CY7,BHQ2 |
(dT-Alexa Flour 568) | R=A,G |
|
5'Alexa Flour 594 |
3'Alexa Flour 594 |
CY7,BHQ3 |
(dT-Alexa Flour 594) | S=G,C |
|
5'Alexa Fluor 610 |
3'Alexa Fluor 610 |
FAM,BHQ1 |
(dT-Alexa Fluor 610) | V=A,G,C |
|
5'Alexa Fluor 633 |
3'Alexa Fluor 633 |
FAM,BHQ2 |
(dT-Alexa Fluor 633) | W=A,T |
|
5'Alexa Flour 647 |
3'Alexa Flour 647 |
FAM,DAB |
(dT-Alexa Flour 647) | Y=C,T |
|
5'Alexa Flour 680 |
3'Alexa Flour 680 |
FAM,ECL |
(dT-Alexa Flour 680) | |
|
5'Alexa Fluor 700 |
3'Alexa Fluor 700 |
FAM,MGB |
(dT-Alexa Fluor 700) | |
|
5'Alexa Fluor 750 |
3'Alexa Fluor 750 |
FAM,TAM |
(dT-Alexa Fluor 750) | |
|
5'ATTO 425 |
3'ATTO 425 |
HEX,BHQ1 |
(dT-ATTO 425) | |
|
5'ATTO 550 |
3'ATTO 550 |
HEX,BHQ1 |
(dT-ATTO 550) | |
|
5'ATTO 565 |
3'ATTO 565 |
HEX,BHQ2 |
(dT-ATTO 565) | |
|
5'ATTO Rho11 |
3'ATTO Rho11 |
HEX,BHQ2 |
(dT-ATTO Rho11) | |
|
5'ATTO 594 |
3'ATTO 594 |
HEX,ECL |
(dT-ATTO 594) | |
|
5'ATTO 655 |
3'ATTO 655 |
HEX,MGB |
(dT-ATTO 655) | |
|
5'ATTO 700 |
3'ATTO 700 |
HEX,TAM |
(dT-ATTO 700) | |
|
5'Acrydite |
3'Azide(N3) |
JOE,BHQ1 |
(dT-Azide(N3) | |
|
5'Azide(N3) |
3'AMCA |
JOE,BHQ2 |
(5F-dU) | |
|
5'BHQ1 |
3'BHQ0 |
JOE,DAB |
(2-F-rA) | |
|
5'BHQ2 |
3'BHQ1 |
JOE,MGB |
(2-F-rC) | |
|
5'BHQ3 |
3'BHQ2 |
JOE,TAM |
(2-F-rG) | |
|
5'BIO |
3'BHQ3 |
JOE.ECL |
(2-F-rU) | |
|
5'BIO-TEG |
3'BBQ-650 |
PHO,FAM |
(2'-O-Methyl -A) | |
|
5'C6-NH2 |
3'BKHFQ (Double Quenching) |
Quasar 670,BHQ3 |
(2'-O-Methyl -C) | |
|
5'C7-NH2 |
3'BIO |
Quasar570,BHQ2 |
(2'-O-Methyl -G) | |
|
5'C12-NH2 |
3'BIO-TEG |
ROX,BHQ2 |
(2'-O-Methyl -U) | |
|
5'CHCH |
3'C3-FAM |
ROX,ECL |
(dT-Digoxin) | |
|
5'CHO |
3'C6-NH2 |
ROX,MGB |
(dT-DNP) | |
|
5'Cholesteryl |
3'C7-NH2 |
TAM,BHQ2 |
(dT-BHQ1) | |
|
5'COOH |
3'CHCH |
TAM,DAB |
(dT-BHQ2) | |
|
5'CY3 |
3'Cholesteryl |
TAM,ECL |
(dT-BHQ3) | |
|
5'CY5 |
3'CY3 |
TAM,MGB |
(dT-Biotin) | |
|
5'CY5-M |
3'CY5 |
TET,BHQ1 |
(dT-Chromeo 494) | |
|
5'CY5.5 |
3'CY5-M |
TET,BHQ2 |
(dT-CY3) | |
|
5'CY7 |
3'CY5.5 |
TET,DAB |
(dT-CY5) | |
|
5'DAB |
3'CY7 |
TET,ECL |
(dT-CY5-M) | |
|
5'DBCO |
3'DAB |
TET,MGB |
(dT-FAM) | |
|
5'DIG |
3'DBCO |
TET,TAM |
(dT-FITC) | |
|
5'ET-ROX |
3'ddC |
TXR,BHQ2 |
(dT-HEX) | |
|
5'ET-TAMRA |
3'DIG |
TXR,DAB |
(dT-Methylene Blue) | |
|
5'ET-B (Blue) |
3'ECL |
TXR,MGB |
(dT-Quasar 570) | |
|
5'ET-G (Green) |
3'Ferrocene |
VIC,BHQ1 |
(dT-Quasar 670) | |
|
5'ET-Y (Yellow) |
3'HEX |
VIC,BHQ2 |
(dT-Quasar 705) | |
|
5'ET-R (Red) |
3'InvdT |
VIC,DAB |
(dT-ROX) | |
|
5'ET-P (Purple) |
3'Joe |
VIC,MGB |
(dT-SF670) | |
|
5'ET-0 (Orange) |
3'MGB |
VIC,TAM |
(dT-TAM) | |
|
5'FAM |
3'Methylene Blue |
|
(dT-VIC) | |
|
5'FITC |
3'PHO |
|
(dT-NH2) | |
|
5'Ferrocene |
3'QSY7 |
|
(HMC) | |
|
5'HEX |
3'ROX |
|
(Me-dC) | |
|
5'HMC |
3'RHO 101 |
|
(PC Linker) | |
|
5'JOE |
3'SF670 |
|
(C7-NH2) | |
|
5'Me-DC |
3'-CT+Signal |
|
(dI) | |
|
5'Methylene Blue |
3'SH |
|
(dU) | |
|
5'NED |
3'Spacer(C3) |
|
(dSpacer) | |
|
5'PHO |
3'SS |
|
(Spacer C3) | |
|
5'Quasar 570 |
3'TAM |
|
(Spacer C6) | |
|
5'Quasar 670 |
3'TXR |
|
(Spacer 9) | |
|
5'Quasar 705 |
3'VIC' |
|
(Spacer C12) | |
|
5'ROX |
|
|
(Spacer 18) | |
|
5'rApp |
|
|
(SS) | |
|
5'SF670 |
|
|
(TAO) | |
|
5'SH |
|
|
(XEN) | |
|
5'SS |
|
|
-CT+Signal | |
|
5'TAM |
|
|
LNA-A | |
|
5'TET |
|
|
LNA-C | |
|
5'TXR |
|
|
LNA-G | |
|
5'triple Biotin |
|
|
LNA-T | |
|
5'VIC |
|
|
(dC-Alexa Flour 350) | |
|
5'YakYel |
|
|
(dC-Alexa Flour 405) | |
|
|
|
|
(dC-Alexa Flour 488) | |
|
|
|
|
(dC-Alexa Flour 532) | |
|
|
|
|
(dC-Alexa Flour 546) | |
|
|
|
|
(dC-Alexa Flour 555) | |
|
|
&nbs |
|||
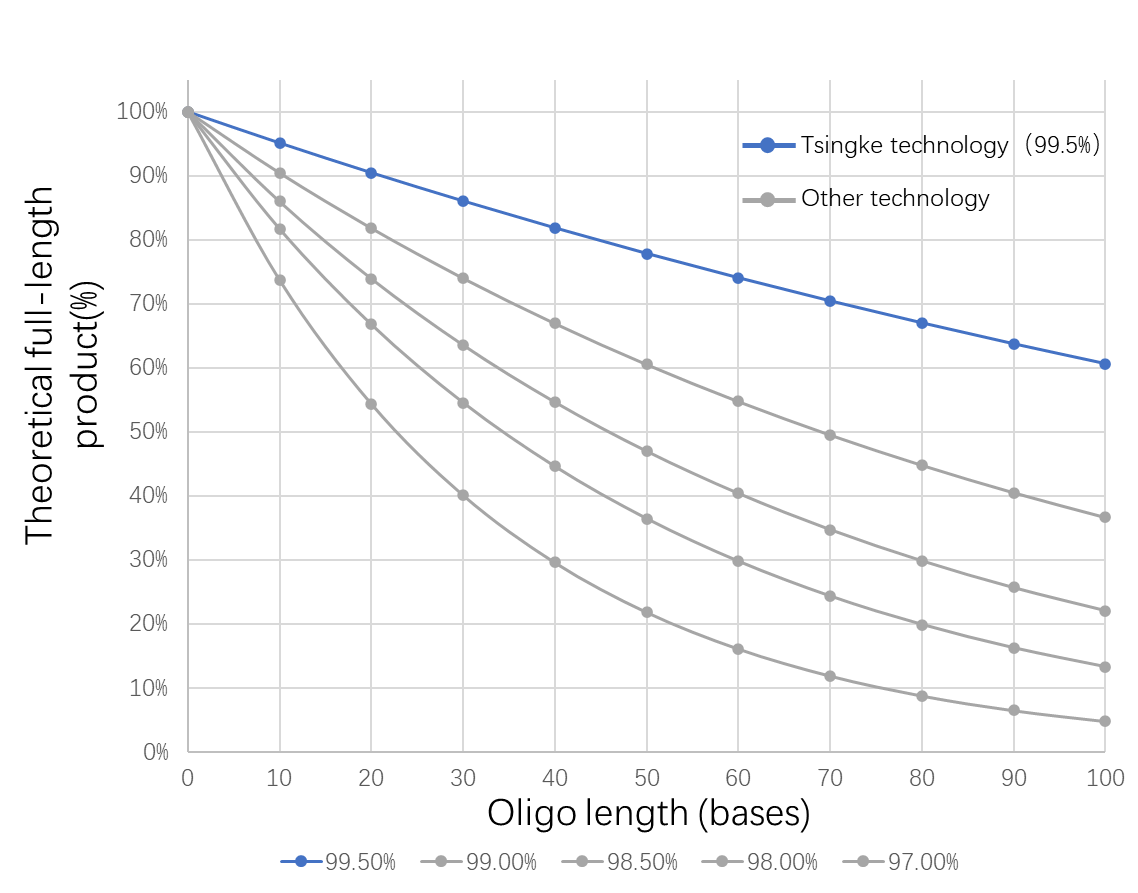
1. Stability and Efficiency: Modified monomers are less stable and have longer coupling times, resulting in lower efficiency and yield.
2. Purification: They require intensive purification methods like PAGE or HPLC, leading to significant material losses.
3. Raw Material Cost: The raw materials for modified primers are much more expensive, often hundreds of times more than those for general primers.